Manolis Kellis
MIT 6.874/6.802/20.390/20.490/HST.506 Spring 2021 Prof. Manolis Kellis
Guest lecture: Wengong Jin
Deep Learning in the Life Sciences / Computational Systems Biology
Playlist: https://youtube.com/playlist?list=PLypiXJdtIca5sxV7aE3-PS9fYX3vUdIOX
Latest slides and course today: http://compbio.mit.edu/6874
Spring 2021 slides and materials: http://mit6874.github.io/
0:00 Introduction
1:16 Drug discovery
3:57 Computational drug discovery
17:14 Deep learning
22:27 Antibiotic discovery
26:30 Traditional approaches
30:16 Antibiotic discovery using GNNs
47:32 Biology aware models
54:35 Incorporating biology and chemistry
1:05:54 De novo drug design
1:06:57 Graph generation
1:13:51 Junction tree variational autoencoder
1:25:03 Conclusion
Source
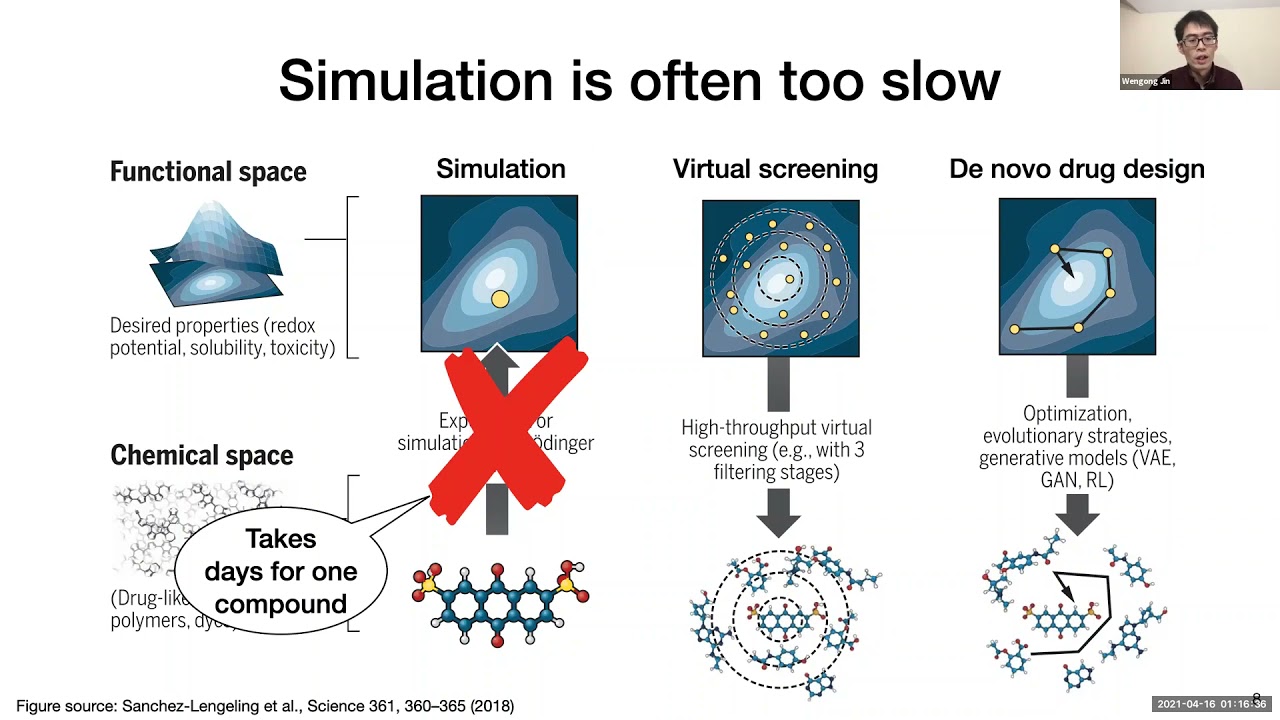


keep it up
Amazing lecture !
great content
Thanks. Super quality.
It's great that lectures like these are publicly available.
Brilliant! Thanks for making this available
life saving lecture omg
Great lecture. For the compound on the right at 47.36min with better MIC, did you observe the different moieties by cleaving the azo bond? It is my guess that the aminosulphonamide or the hydrazine analog may be the actual active drug and not the whole compound shown.
is ComboNet publicly accessible?
Ambitious I like…🤓
Great lecture! Thank you very much for uploading this. I just got two questions:
1. We know that drugs are 3D when they bind to their targes. But the grape in GNN is a 2D representation of drug structures. For some drugs which are only active in one enantiomer form not the other one, the 2D graph will be the same for the two enantiomers. How can GNN capture this stereochemical difference?
2. Remdesivir used in the ComboNet on slide 36 is a prodrug, its active form is a metabolite which is a triphosphate. I suppose most of the drugs used in the training are not prodrug, so I'm curious about whether remdesivir or it's active form is used in the work? Did the author consider this?
why look for a cv19 drug? There are so many more deadly conditions to treat that need drug discovery!
Thank you. The lecture is great.
This is an excellent resource for getting to know GNNs in drug de novo drug design, thank you!
Thanks for this lecture. I have watched this like three time today just to capture the mind behind the presentation. please, do you have a github for the work? while I understand the VAE and GNN. I don't understand how you created the junction tree VAE.
Also, does encoding of just one molecule unto a low dimension lead to the vast chemical space?
can one introduce a sort of motif library e.g. ligand library for developing new Transition metal complex with desired properties like catalytic activity?
Robotized measurement —> https://www.youtube.com/watch?v=dEW5YO7l5TQ
For more details see —> https://www.nature.com/articles/s43246-022-00235-5
An amazing lecture indeed. I just wanted to clarify as to how the viral/host targets are represented or vector encoded in the combonet model.
2.6B only when considering the failures, so for a startup looking to raise money, its not as important. Rather the average cost to get one drug into FIH trials is more relevant, and then how much to get the $ to run the phase 1 trial. The 2.6B is more a big pharma number.
very helpful! great lecture!
thank you so much for sharing this valuable lecture about GNNs in drug discovery. The idea of junction tree VAE is awesome and making sense
COVID-19 is spread all the world now, so the data might be even more than antibolic🤔
Awesome lecture!
Thank you for sharing this!
I'm sorry Dave, I'm afraid I can't do that.